Welcome to the September 2024 edition of Phytogen!
Welcome Spring! As we are approaching the warmer months, our ASPS conference is getting closer (To register and submit an abstract for your local ASPS hybrid conference HERE).
This issue we congratulate the 2024 ASPS winners and publication by our members. We have a contribution from Pr. Lucas Cernusak, our Education Representative, and a research contribution from Dr. Francine Perrine-Walker. Also included in this issue is 2023 R.N Robertson Travelling Fellowship report. We also continue to highlight our student awardees of the travel grant of the International Plant and Molecular Biology (IPMB) conference held in Cairns in June.
There are several opportunities to be part of ASPS.
We welcome contributions from students and researcher at any time for publication in Phytogen. Its free and a great way to give your research profile a boost and share your findings! Send your contribution to our editors Razlin (razlin.azman@csiro.au) or Lucas (L.Auroux@latrobe.edu.au).
Celebrating achievements
Congratulations to the winners of the 2024 ASPS award who will be presenting at the ASPS hybrid conference happening in late November.

Peter Goldacre Award: Dr Peter Crisp (UQ profile | X: pete_crisp)
Jan Anderson Award: Assoc Prof Jenny Mortimer (UoA profile | X: Jenny_Mortimer1 )
ASPS Education and Outreach Award: Dr Ashley Jones (ANU profile |X: dnawhisperer ) & Assoc Prof Benjamin Schwessinger (ANU profile | X: @schwessinger | Research Group)
JG Wood Lecturer: Prof Sergey Shabala (UWA profile)
In addition, congratulations to ASPS member Dr Lim Chee Liew and colleagues (LaTrobe Uni) for their publication in Nature Plants and the front cover feature.
Image by Lucas Auroux and Lim Chee Lim
Be part of ASPS!
Nominations are now accepted for the following positions in ASPS. Please reach out to Janet Wheeler (Janet.Wheeler@latrobe.edu.au) if you are interested in getting involved in these roles.
- ASPS secretary
- Education and Outreach
- Diversity and Inclusion
- Student representative
Discipline leads:
- Cell Biology Representative
- Plant-Microbe Interactions Representative
Education Representative Update
 Pr. Lucas Cernusak, College of Science and Engineering, James Cook University, Cairns Campus (Queensland)
Pr. Lucas Cernusak, College of Science and Engineering, James Cook University, Cairns Campus (Queensland)
Bio: Pr. Lucas Cernusak | James Cook University
Lucas Cernusak is a Professor of Plant Science at James Cook University, Cairns. His main research interest is to understand the environmental and biological controls on carbon dioxide and water vapour exchange between leaves and the atmosphere. He is also interested in improving the interpretation of stable isotope signals in plant organic material, to better understand how leaf gas exchange has responded to global climate change through time and how it varies across ecological gradients. He and his lab group at James Cook University are especially interested in understanding these processes in tropical trees and responses of tropical rainforests to climate change and rising atmospheric CO2.
Lucas joined James Cook university in 2013 as an ARC Future Fellow, after having completed his PhD in the Research School of Biology at the Australian National University, after which he did postdoctoral research at the Smithsonian Tropical Research Institute in the Republic of Panama and Charles Darwin University in Darwin, Northern Territory. He has been a member of the ASPS since beginning his PhD in 2000. Over his career, Lucas has published more than 150 peer-reviewed journal articles and book chapters.
In addition to his research, Lucas is also passionate about plant science education, outreach, and engagement. Lucas has made substantial contributions to the academic community through mentorship and teaching. He has supervised numerous PhD students to completion, helping his students win research grants and publish their research under his guidance. His dedication to teaching is reflected in exemplary student feedback scores, and he continually strives to make a lasting impact in the classroom.
Looking ahead, Lucas is excited to have been elected to the role of Plant Science Education Representative in the ASPS, and to have the opportunity to promote plant science education in Australia and internationally. He strongly believes that the ASPS provides an ideal vehicle for doing this.
R.N Travel Fellowship Report
Exploring Genome Structure in Cicer echinospermum through Oligo-FISH Chromosome Barcoding

Report by: Alistair Hockey
PhD candidate
The UWA School of Agriculture and Environment, University of Western Australia
In a world of increasing digitalisation, it’s easy to lose touch with the physical components that make up our world. An ongoing reliance on bioinformatic analysis in the fields of genetics and genomics has led to the inference of genetic factors and effects with little to no independent physical evidence. Such is the case with the study of genome structure diversity, a field founded in cytogenetics, now dominated by genome sequence analysis and inference. Genome structure is a key determinant of gene flow dynamics within and between species, with differences in structure often resulting in barriers to gene flow. With the support of the R.N. Robertson Travelling Fellowship, I set out to identify, validate and characterise genome structure diversity in chickpea (Cicer arietinum ssp. arietinum L.) and Cicer echinospermum P.H. Davis through, oligo-FISH chromosome barcoding, a modern cytogenetic approach.

Image: Dolní náměstí, Olomouc city centre
My PhD project is focused on uncovering post-zygotic barriers to gene flow in Cicer echinospermum to better understand the species coherence and relationship to closely related sympatric species. Cicer echinospermum is the only known annual Cicer species (2n = 2x = 16) capable of hybridizing with chickpea to produce semi-fertile hybrids. Moreover, the genetic diversity within C. echinospermum far exceeds that of domesticated chickpea (108 times greater!) (von Wettberg et al., 2018). Introducing novel genetic material to increase genome-wide diversity in chickpea could be beneficial for both short-term and long-term crop improvement. However, barriers to gene flow between the two species exist, large genome structure variation chief among them. Karyotypic variation between the two species was first discovered by Ladizinsky and Adler (1976), who observed meiotic chromosome pairing aberrations in F1 hybrids and concluded the two species differed by a reciprocal translocation. Since then, most identification and characterization of large genome structural variations (> 1 Mbp) between the two species have been through bioinformatic analyses (e.g., pangenomes).
In my PhD project, I am using both bioinformatic and cytogenetic approaches to identify, validate, and characterize genome structure variation within C. echinospermum. Olomouc, a Czech university city in the heart of Moravia, is known for its baroque-style architecture and is home to the Institute of Experimental Botany (UEB). The Hribova Lab at UEB developed an oligo-FISH barcoding system for chickpea. This modern cytogenetic approach allows for the physical characterization of karyotypic diversity in chickpea. Thanks to a collaboration between my primary supervisor Dr. Judith Lichtenzveig and Prof. Jaroslav Dolezel, I had the opportunity to spend June 2024 working with the Hribova Lab in Olomouc to test the chickpea barcoding system on C. echinospermum. Additionally, I attended the EMBO Plant Genome Stability and Change 2024 workshop in Olomouc and presented a seminar on my current PhD findings.
We planned two experiments to produce mitotic chromosome spreads from meristematic root tips and label chromosomes through oligo-FISH barcoding. One experiment involved cell cycle synchronization to prepare mitotic chromosomes at metaphase I, while the other used a simpler protocol to halt cell cycle continuation in meristematic root tips. Both experiments then followed the same protocols to prepare protoplast suspensions and perform fluorescent in situ hybridization. We started with one chickpea variety and one C. echinospermum accession to confirm the reciprocal translocation identified almost 50 years ago. Initially, everything went smoothly, and we clearly identified the reciprocal translocation between the two species based on the oligo-FISH barcodes on chromosomes. We also confirmed that the cell cycle synchronization protocol was more effective for detecting mitotic chromosomes at metaphase I and the protocol worked better for chickpea, the species it was designed.

Image: Probe hybridisation on mitotic spread slides
Subsequent iterations of these experiments faced challenges due to equipment malfunctions and seed germination issues. Additionally, some protocols optimized for chickpea were not optimal for C. echinospermum, which limited probe visualization for several C. echinospermum accessions. In response, we prepared for future experiments at the University of Western Australia, where I can use the expertise I gained to optimise the protoplast suspension protocol and perform oligo-FISH barcoding to investigate genome structure diversity within C.echinospermum.

Image: Plant Genome Stability and Change, 2024
My time at the EMBO workshop was invaluable. Presenting my research sparked discussions with peers and leaders in my field. The exposure to innovative and intriguing research helped me develop ideas for future projects and opened my eyes to the opportunities available. It was fantastic to just spend four days discussing all of the new research in great company. All in all, a month spent in the European summer is never wasted. After long days in the lab, I often walked the perimeter of the old city, following the parklands that once formed a moat around the medieval fortifications ‒ a daily blend of the modern and the ancient. I can’t imagine a better place to have further developed my abilities in plant cytogenetics. The welcoming nature of everyone at UEB immediately made me feel at home, especially after a sunny afternoon roasting špekáček sausages over an open fire on the institute grounds, paired with Czech pilsner.
I am particularly grateful to Jana Cizkova and Eva Hribova, whose attentiveness, interest, and openness provided me with an unparalleled learning opportunity. Their introductions to research leaders from other labs at UEB sparked discussions that gave me novel insights and ideas for future research. I’d also like to thank my supervisor Judith Lichtenzveig, whose inspiration for the research trip, involvement in approach development, and ongoing supervision of this research project cannot be understated. Lastly, I thank the ASPS for awarding me the R.N. Robertson Travelling Fellowship, which allowed this research trip to take place, and the University of Western Australia’s Graduate Research School for additional funding.
Research Spotlight
Bacterial Biosensors: The cumate-inducible sfGFP expression system to study Rhizobium leguminosarum bv viciae – vetch interaction
Written by: Francine Perrine-Walker (PhD), The University of Sydney (LinkedIn | Google Scholar | X: PerrineWalker)
There have been some advances in the use biosensors in rhizobia to monitor oxygen and proline in Pisum sativum nodules [1,2], root exudates in the pea and vetch rhizosphere [3] and glutamine in the alfalfa, lentil, pea and soybean roots and nodules [4]. So, imagine if you could have a microscopic tool in your lab such as a Rhizobium bacterial cell sensing any compound released by a legume or a non-legume plant and monitor how such cells reacted in the rhizosphere or within roots.
In molecular biology, inducible gene expression systems (or vectors) have been useful tools for biotechnological applications. In such systems, an inducer, when present allosterically, binds to and renders inactive a transcriptional regulator/repressor which is bound to the operator sites in the promoter region of target genes [5]. Inducers can be anything such as glycerol [6], anthranilate [7], or even green light [8]. Kaczmarczyk and colleagues [5] demonstrated the advantages of inducible gene expression systems from Pseudomonas putida [9, 10] that could be induced in the presence of p-cumate and synthetic vanillate in sphingomonads and Alphaproteobacteria. Interestingly, Assoc. Prof. N. Coleman and M. Sommerville (University of Sydney) had a Rhizobium strain, R. leguminosarum bv. viciae 3841, in their collection expressing the cumate-inducible expression system i.e., the plasmid pUS248-sfGFP (superfolder green fluorescence protein).
To test this if the cumate-inducible expression system in R. leguminosarum bv. viciae 3841 could be used to monitor infection and nodulation of legumes in the presence of p-cumate, Namoi woolly pod vetch was used in two different growth conditions: a) the mounted glass-slide growth system previously described in Fahräeus [11] and modified in Perrine-Walker et al. [12] and, b) the standard nodulation assays [11, 13]. As inoculants, the wild-type R. leguminosarum bv. viciae 3841 (WT) and the transformant, R. leguminosarum bv. viciae 3841 expressing the pUS248-sfGFP were grown on YMA [14] and YMA agar plates supplemented with kanamycin at 25 µg/mL (Km 25) to maintain the pUS248-sfGFP in transformants and p-cumate at 50 mM to activate GFP expression at 30oC respectively. Plant materials (intact roots and/or sectioned, squashed nodules) were examined using fluorescence microscopy.
Preliminary results were encouraging. For the mounted glass-slide growth system, in the presence of p-cumate, GFP-expressing R. leguminosarum bv. viciae 3841 pUS248-sfGFP transformants adhered/attached to the surface of a root hair post 90 min inoculation (Fig. 1 c). At 24, 48 and 72 h, the fluorescent bacteria were found trapped in curled root hairs and within infected cortical cell regions below infected root hairs (Fig. 1 d-j) compared to control uninoculated vetch seedlings (Fig. 1 b). A GFP signal was observed in sectioned roots i.e., within infection threads traversing cortical cell region (Fig. 1 h-j).
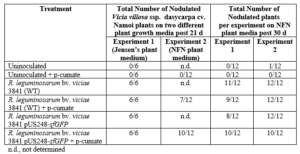
For nodulation assays, no nodules were observed on the roots of uninoculated plants, whereas nodules were observed in WT strain inoculated plants post 21 and 30 d (Table 1). Furthermore, no or diffuse GFP signal was detected within infected cells of sectioned nodules of inoculated p-cumate treated R. leguminosarum bv. viciae 3841 pUS248-sfGFP-inoculated plants (Fig. 1 k-m). In NFN-medium, with and without p-cumate, no nodules were observed on control plants. Plants inoculated with WT strain formed nodules (Table 1) however in the presence of p-cumate, no GFP signal was observed in squashed nodules or in root sections under the microscope after 21 and 30 d post inoculation. In plants inoculated with R. leguminosarum bv. viciae 3841 pUS248-sfGFP in the absence of p-cumate, nodulation occurred (Tables 1) but no GFP signal was observed in nodules and on the surface of root sections after 21 and 30 d post inoculation. In the presence of p-cumate, GFP signals were observed on the surface of root sections, in infection threads found within curled root hairs that extended towards putative nodule primordia (Fig 1. n) post 21d inoculation and zone II section of vetch nodules on seedlings inoculated with R. leguminosarum bv. viciae 3841 pUS248-sfGFP post 30 d inoculation (Fig 1. p-o).
In conclusion, the cumate-inducible gene expression system did not affect root hair attachment and curling, infection thread infection and nodulation of the Rhizobium leguminosarum bv. viciae strain 3841 with the plasmid pUS248-sfGFP (superfolder green fluorescence protein) in the presence of p-cumate (summarized in Figure 2). The inducible expression vector with the superfolder GFP could be useful, but more work would be needed to determine if the construct could be modified to be inducible by another compound other than p-cumate.
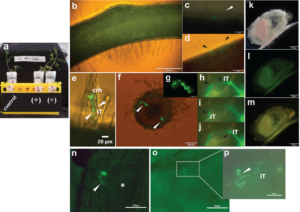
Fig. 1 Visualization of Rhizobium leguminosarum bv. viciae 3841 pUS248-sfGFP in vetch roots using the mounted glass slide method and nodulation assays; (a) Growth of individual vetch plants uninoculated (control) and inoculated with R. leguminosarum bv. viciae 3841 pUS248-sfGFP using the mounted glass slide method in the presence of Kanamycin at 25 µg/mL and p-cumate at 50 mM (+); (b) uninoculated vetch root (control); (c) GFP- labelled bacteria on the surface of an individual root hair post 90 min inoculation (white arrowhead); (d) root hair deformation (black arrowheads) and sfGFP-expressing rhizobia on the plant root surface; (e) sfGFP-labelled bacteria within two putative developing infection threads in a curled root hair (white arrow); (f) two infection sites within the cortical cell region of a sectioned vetch root (white arrowheads); (g) close-up of one the infection sites shown in (f); (h-j) close up of the infection site in (f) showing sfGFP-labelled rhizobia within the infection thread traversing three cortical cells; (k-m) hand-sectioned nitrogen-fixing nodule on a vetch root inoculated R. leguminosarum bv. viciae 3841 pUS248-sfGFP; (k) under bright field; (l) same nodule in (k) under the U-MGFPHQ filter specific for GFP; (m) same nodule in (k) under U-MWIB2 filter showing diffuse GFP fluorescence signals; (m)infection threads containing GFP-labelled rhizobia growing towards a putative nodule primordium (white asterisk); (n) infection thread junction with two sfGFP-labelled bacteria within a zone II nodule region; (o) squashed zone II region of a vetch nodule displaying infection threads with and without GFP signals and; (p) close up of an infection thread section in (o; white dashed rectangle) containing sfGFP-labelled rhizobia. Note – Images in (h-j) are the same region at three different focal planes to highlight the infection thread (IT). Fluorescence images (b), (c), (d), (e) and (f) were captured using U-MWIB2 filter with bright field light (h-j) and (m) with the same filter with no bright field light. Images in (g) and (l) were captured under the U-MGFPHQ filter specific for GFP only. (b) to (m) were captured using the Olympus BX51 microscope and seedlings were grown on Jensens plant media supplemented with p-cumate. (n) to (p) were captured using the Nikon Ti2 microscope and seedlings were grown on NFM supplemented with p-cumate. crh, curled root hair; IT, infection thread.
Table 1. Nodulation of Namoi woolly pod vetch by R. leguminosarum bv. viciae strain 3841 and R. leguminosarum bv. viciae 3841 pUS248-sfGFP post 21 d on Jensen’s and NFM plant growth medium without kanamycin. Each experiment had a total of 6 and 12 plants for each treatment respectively. The number of plants that nodulated were recorded post 21 or 30 d.
![]()
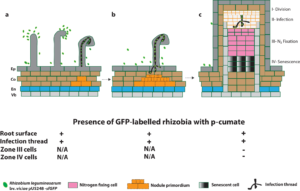
Fig. 2 Schematic diagram summarizing the expression of sfGFP in rhizobia in vetch in the presence of p-cumate in plant growth media. In the presence of p-cumate in plant growth media, GFP signal were observed on surface of vetch roots as well as to the tips of root hairs and curled root hairs and within infection threads (a). (b) sfGFP-labelled rhizobia were also in infection threads growing towards nodule primordia and (c) infection threads with the zone II of vetch nodules. No GFP signal were found in zone III and zone IV of vetch nodules.
Acknowledgments
I would like to thank Dr J. Hartley for kindly donating the vetch seeds and M. Sommerville for the Rhizobium strains and helpful advice as well as Profs. B. Kaiser and D. Guest for access to laboratory materials and fluorescent microscopes. I would also like to thank Assoc. Prof. N. Coleman for use of the Rhizobium strain expressing the plasmid pUS248-sfGFP. Plasmid pUS248-sfGFP (a gift from Nicholas Coleman’s lab, University of Sydney) was made by joining the replication and resistance regions of pBBR1MCS-2 [15] to the p-cumate regulatory system components derived from Pseudomonas putida [5] and the superfolder GFP gene (sfGFP) [16].
References
- Rutten PJ, Steel H, Hood GA, Ramachandran VK, McMurtry L, Geddes B, Papachristodoulou A, Poole PS (2021) Multiple sensors provide spatiotemporal oxygen regulation of gene expression in a Rhizobium-legume symbiosis. PLOS Genetics 17 :e1009099. doi:10.1371/journal.pgen.1009099
- Rubia MI, Ramachandran VK, Arrese-Igor C, Larrainzar E, Poole PS (2020) A novel biosensor to monitor proline in pea root exudates and nodules under osmotic stress and recovery. Plant Soil 452 :413-422
- Pini F, East AK, Appia-Ayme C, Tomek J, Karunakaran R, Mendoza-Suarez M, Edwards A, Terpolilli JJ, Roworth J, Downie JA, Poolea PS (2017) Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol 174 :1289-1306
- Thilakarathna MS, Raizada MN (2018) Visualizing glutamine accumulation in root systems involved in the legume–rhizobia symbiosis by placement on agar embedded with companion biosensor cells. Phytobiomes Journal 2 :117-128
- Kaczmarczyk A, Vorholt JA, Francez-Charlot A (2013) Cumate-inducible gene expression system for sphingomonads and other alphaproteobacteria. Appl Environ Microb 79 :6795-6802
- Doi Y (2015) L-lactate production from biodiesel-derived crude glycerol by metabolically engineered Enterococcus faecalis: Cytotoxic evaluation of biodiesel waste and development of a glycerol-inducible gene expression system. Appl Environ Microb 81 :2082-2089
- Hoffmann L, Sugue MF, Bruser T (2021) A tunable anthranilate-inducible gene expression system for Pseudomonas Appl Microbiol Biot 105:247-258
- Abe K, Miyake K, Nakamura M, Kojima K, Ferri S, Ikebukuro K, Sode K (2014) Engineering of a green-light inducible gene expression system in Synechocystis PCC6803. Microb Biotechnol 7:177-183
- Eaton RW (1996) p-Cumate catabolic pathway in Pseudomonas putida F1: Cloning and characterization of DNA carrying the cmt operon. J Bacteriol 178 :1351-1362
- Eaton RW (1997) p-Cymene catabolic pathway in Pseudomonas putida F1: Cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol 179 :3171-3180
- Fahräeus G (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16 :374-381
- Perrine-Walker FM, Kouchi H, Ridge RW (2014) Endoplasmic reticulum-targeted GFP reveals ER remodeling in Mesorhizobium-treated Lotus japonicus root hairs during root hair curling and infection thread formation. Protoplasma 251 :817-826
- Jensen HL (1942) Nitrogen fixation in leguminous plants. II. Is symbiotic nitrogen fixation influenced by Azotobacter? Proc Linn Soc NSW 67: 205-212
- Allen EK, Allen ON (1950) Biochemical and symbiotic properties of the rhizobia. Bacteriol Rev 14 :273-330
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166 :175-176
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24 :79-88
Student travel grant awardees report – IBMP Cairns 2024
 Samuel James Nix – ANU (ACT)
Samuel James Nix – ANU (ACT)
Samuel Nix | ANU Research School of Biology
Overall, the conference was a fantastic experience. There were multiple plenary talks that I enjoyed. Robert Furbank’s C4 Rice Project presentation was a good presentation on how difficult multi-gene transgenic projects can be approached. It was also interesting to see how biotechnology has advanced in the past 15 years and how that impacted the feasibility of the project. Another talk I enjoyed was Mohammad Ishiyaku’s presentation on GM Cow Pea, a great success story about how a GM crop can be embraced by a developing country and the local communities and be adapted to meet the commercial desires of the public. A fun plenary talk was Susana Coelho’s on sex evolution in brown algae. Additionally, I presented my poster Elucidating and Engineering Cell Specific Cyclic Electron Flow in C4 photosynthesis and I won an award! Lastly, and most importantly, I made new connections with fellow scientists during the conference.
 Sara Jalali – UoA (SA)
Sara Jalali – UoA (SA)
Sara Jalali | Researcher Profiles (adelaide.edu.au)
I, Sara Jalali, had the privilege of attending the International Plant Molecular Biology (IPMB) Conference in Cairns, Australia, from June 24th to June 28th, 2024. This conference was an invaluable experience, allowing me to engage with leading researchers in molecular biology, a field that directly relates to my PhD work. The event facilitated the exchange of innovative ideas and the discussion of potential collaborative research opportunities. Additionally, it helped me expand my professional network and stay informed about the latest advancements in plant molecular biology. I am deeply grateful to the ASPS for funding my travel, enabling me to participate in this enriching event.
 Rana Alqusumi – UWA (WA)
Rana Alqusumi – UWA (WA)
Profile: Rana Alqusumi (plantenergy.edu.au)
At the end of June 2024, I attended the IPMB 2024 Congress in Cairns, thanks to the ASPS travel award. The event prominently featured emerging researchers in plant molecular biology. It was an honour to listen to scientists from around the world discuss their latest advancements in plant science. One of the highlights was a presentation by Prof. Mitter from the University of Queensland, who shared her team’s research on using clay nanosheets for delivering RNAi as pesticides. This innovative application of nanotechnology in crop protection was particularly enlightening. Additionally, I had the pleasure of engaging with fellow plant researchers, exchanging insights about our respective fields of work. The congress provided ample opportunities for networking and collaboration. Beyond the academic experience, I also enjoyed exploring the city of Cairns. Overall, attending the IPMB 2024 Congress was an enriching experience offering valuable knowledge and memorable interactions.
 Saber Sohrabi – UoA (SA)
Saber Sohrabi – UoA (SA)
Saber Sohrabi | Researcher Profiles (adelaide.edu.au)
I, Saber Sohrabi, attended the International Plant Molecular Biology (IPMB) Conference, which took place from June 24th to June 28th, 2024, in Cairns, Australia. By participating in this conference, I had the chance to connect with researchers in my field of study, exchange ideas, and explore potential collaborations for future research projects. The conference focused on molecular biology, aligning perfectly with my PhD project. This experience allowed me to expand my professional network, stay updated on the latest developments, and establish myself as an active member of the academic community. I would like to thank the ASPS for funding my flights to attend this conference.
 Alicia Quinn – Monash (VIC)
Alicia Quinn – Monash (VIC)
LinkedIn | Twitter AliciaQuinnSci
Attending the IPMB 2024 conference in Cairns was a great opportunity to see the latest research in plant molecular biology and meet the people behind the work. The conference started strong with two excellent plenaries that set the tone for the rest of the week: Gerry Turpin on protecting Indigenous knowledge and Tetsuya Higashiyama on signalling during reproduction. The concurrent sessions were equally diverse – covering everything from evolution to cell signalling – and it was a challenge deciding what to attend. Presenting my work in the metabolic engineering session was a highlight, and the supportive audience made it a fantastic experience. I really enjoyed the evening poster sessions and it was great to chat about the exciting research currently underway. This conference was an invaluable experience and I’m grateful for the support from ASPS to attend. I look forward to the next one!
Editors’ note
Phytogen will be back for our next issue in late October Don’t forget, registration and abstract submission for the local ASPS meetings for each state/territory in Australia and New-Zealand are now open!
For those working outdoors, remember to stay protected, cool and hydrated out there.

