- About
- Members
- Join
- Member log in
- Membership Renewal
- Member directory
- Life Members
- ASPS Life Member Professor Graham Farquhar
- ASPS Life Member Associate Professor Hendrik (Hank) Greenway
- ASPS Life Member Dr Marshall (Hal) D Hatch
- ASPS Life Member Dr Paul E Kriedmann
- ASPS Life Member Dr Mervyn Ludlow
- ASPS Life Member Emeritus Professor Rana Munns
- ASPS Life Member Conjoint Professor Christina E Offler
- ASPS Life Member Professor (Charles) Barry Osmond
- ASPS Life Member Emeritus Professor John W Patrick
- ASPS Life Member Dr Joe Wiskich
- Corresponding Members
- Elected Fellows
- Events
- Awards & Funding
- Employment
- Publications
- Research
- Teaching
- Menu
May Phytogen out now. GPC and STA newsletters
31 May 2023
Hello ASPS members,
The May edition of Phytogen is out now and can be accessed HERE.
The GPC e-bulletin can be accessed HERE for May edition, HERE for April edition.
STA newsletters can be accessed HERE
31 May 2023
Hello ASPS members,
The May edition of Phytogen is out now and can be accessed HERE.
The GPC e-bulletin can be accessed HERE for May edition, HERE for April edition.
STA newsletters can be accessed HERE
May 2023 Phytogen
28 May 2023
Welcome to Phytogen for May 2023. Soon registration will open for ASPS2023 in Tasmania in November Tuesday 28th November to Friday 1st December. Details so far are available at https://www.asps.org.au/conferences/asps-2023
To get you ready for preparing your abstacts, here are reports from poster prize winners in 2022. We hope you enjoy reading:
Alyssa Martino, PhD Candidate, School of Life and Environmental Sciences, The University of Sydney.
What is the title of your poster? 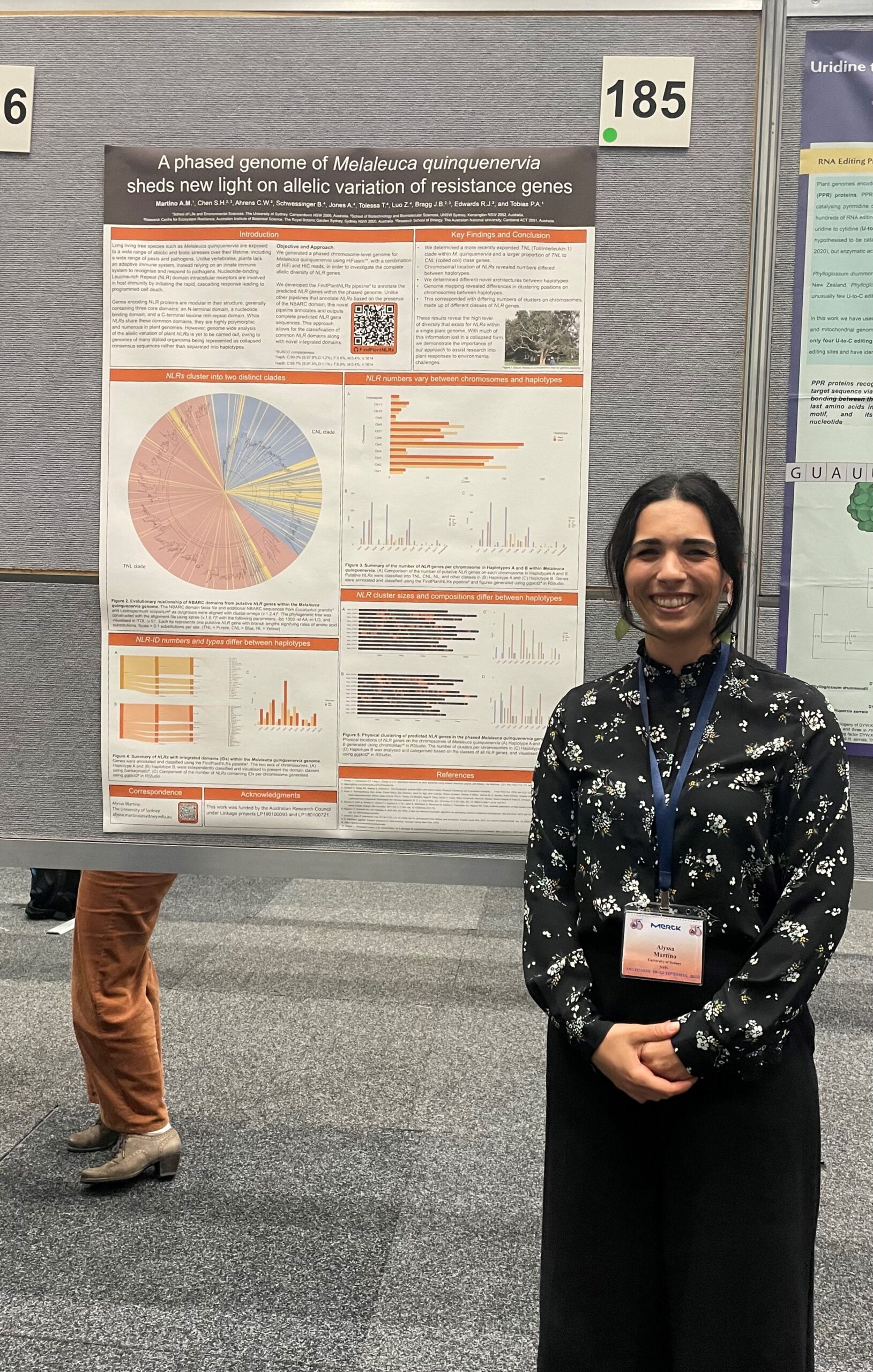
A phased genome of Melaleuca quinquenervia sheds new light on allelic variation of resistance genes
What is the aim of your research, and what is the main hypothesis that you are testing?
Long living tree species are exposed to a wide range of abiotic and biotic stresses in their lifetime, including numerous pests and pathogens. Unlike animals, they lack an adaptive immune system, instead relying on an innate immune system to recognise and respond to pathogens. Plant Nucleotide-binding Leucine-rich repeats (NLRs) are modular intracellular receptors involved in host immunity by initiating the rapid, cascading response leading to programmed cell death. Genes encoding NLRs are highly polymorphic and numerous in plant genomes. However, genome wide analysis of the allelic variation of plant NLRs is yet to be carried out, owing to genomes of many diploid organisms being presented as collapsed consensus sequences rather than separated into haplotypes. Therefore, the focus of this project was to investigate the allelic diversity of NLRs in a long-living tree species.
What plant species was the focus of your research, and what were the key techniques used?
We generated a phased chromosome-level genome for the long-living tree species Melaleuca quinquenervia (broadleaf paperbark). Using HiFiasm software with a combination of HiFi and HiC data, the genome was phased into haplotypes to investigate the complete allelic diversity of NLR genes. The FindPlantNLRs pipeline was developed to annotate the predicted NLR genes within the phased genome, including novel integrated domains.
What was the major finding(s) of your research?
Genome mapping revealed differences in the clustering position of NLRs on chromosomes between haplotypes, which corresponded with differing numbers of NLR clusters between haplotypes. Different novel domain architectures were also detected between haplotypes, demonstrating a high level of haplotype diversity of NLR genes within the M. quinquenervia genome. With much of this information lost in a collapsed form, we demonstrate the importance of our approach to assist research into plant responses to environmental challenges.
Who were the important collaborators and funders of your work?
The work was carried out in collaboration with Stephanie Chen (UNSW), Collin Ahrens (UNSW), Benjamin Schwessinger (ANU), Ashley Jones (ANU), Tamene Tolessa (ANU), Zhenyan Luo (ANU), Jason Bragg (UNSW), Richard Edwards (UWA), and Peri Tobias (USyd) and was funded by ARC Linkage projects LP190100093 and LP180100721 and Research Training Program Stipend Scholarships (Alyssa and Stephanie).

Zheng (Tommy) Gong, HDR Student, PGEL|School of Agriculture and Food Science, The University of Queensland.
What is the title of your poster?
A simple and rapid workflow to evaluate CRISPR gRNA genome editing efficiency in planta.
What is the aim of your research, and what is the main hypothesis that you are testing?
This research aims to develop a workflow that would allow researchers to easily test the efficiency of their selected CRISPR gRNA in planta targeting any sequence-of-interest.
What plant species was the focus of your research, and what were the key techniques used?
We used Nicotiana benthamiana, a model plant species, for transient expression of our CRISPR cassettes. To increase the applicability of this workflow, only general molecular biology skills were required to evaluate the efficiency of multiple gRNAs.
What was the major finding(s) of your research?
We first conceptualised the workflow to be as simple as possible with minimal molecular cloning required. Using this workflow, we successfully evaluated the efficiency of multiple gRNAs targeting an endogenous N. benthamiana gene. We found that the in planta genome editing efficiency of gRNAs was not consistent with that predicted by in silico gRNA design tools, demonstrating the importance of a workflow as such. Using this workflow, we have demonstrated and intend to in future studies, evaluate gRNAs targeting sequences in crop species and other agriculturally important plants.
Who were the important collaborators and funders of your work?
Important collaborators of this work are Yijie Wang and Genevieve Durrington who are currently PhD students at The University of Queensland and my excellent supervisors, Prof. Jimmy Botella, and Dr Karen Massel.
I am funded by the Australian Government RTP scholarship.
Nick Booth, College of Science & Engineering, Flinders University of South Australia.
Nodulin 26 is a multifunctional plant aquaporin that facilitates ammonium transport in nitrogen-fixing soybean nodules.
Booth N.1, Ramesh S.1, Jenkins C.1, Soole K.1, Tyerman S.2, and Day D.1
1College of Science & Engineering, Flinders University, GPO Box 5100, Adelaide, SA 5001, Australia.
2ARC Centre of Excellence in Plant Energy Biology, School of Agriculture, Food and Wine, University of Adelaide, Glen Osmond, South Australia 5064, Australia.
At COMBIO 2022 I was awarded an ASPS poster prize for presenting our findings on the soybean aquaporin, Nodulin 26 (NOD26). Legumes have developed a symbiotic relationship with rhizobia, a soil bacterium that fixes atmospheric nitrogen into a form usable by the plant. During the symbiosis, legumes develop highly specialised organs on their roots, called nodules, which are infected by rhizobia. Inside the infected nodule cells, the rhizobia differentiate into nitrogen-fixing bacteroids that are excluded from the plant cytosol in organelle-like structures termed symbiosomes. The symbiosome membrane acts to regulate the metabolite exchange between the symbionts. Interestingly, a non-selective cation channel which facilitates the efflux of fixed nitrogen from symbiosomes remains unidentified (Tyerman et al. 1995). We proposed that NOD26 may be this non-selective cation channel and begin to characterise the channel through two-electrode voltage clamp in Xenopus laevis oocytes, thanks to a collaboration with Stephen Tyerman, Adelaide University.
Ion transport through aquaporins has long been controversial, with Yool and colleagues (1996) first reporting this in human aquaporin 1. More recently, non-selective cation activity has been reported in Arabidopsis PIP2;1, which shares several characteristics with the soybean symbiosome membrane non-selective cation channel. Our work has shown that when NOD26, a major component of the soybean symbiosome membrane, is heterologously expressed in X. laevis oocytes, monovalent cation-induced currents are observed. The channel is permeable to ammonium, methylammonium, potassium and sodium but not choline or anions, and that these currents are inhibited by divalent cations. Through phosphomimetics of S262, we identified a switch in the permeability of NOD26 to water or cations when expressed in X. laevis oocytes (Figure 1). We believe that the soybean aquaporin NOD26 is multifunctional channel that facilitates both ammonia and ammonium transport in nitrogen-fixing nodules and that this transport is regulated by phosphorylation of S262.
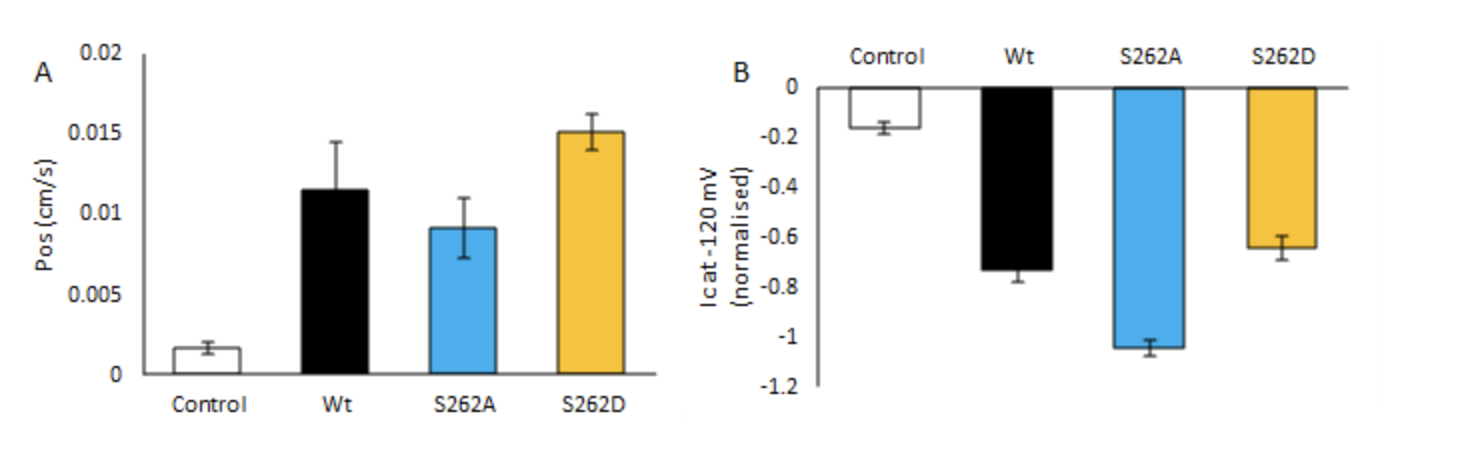 Figure 1. Mutations to GmNOD26 alter water and cation permeability. (A) Osmotic water permeability; or (B) Na+ induced clamp currents; of control, Phosphomimetics or Wt GmNOD26-expressing X. laevis oocytes (n = 8).
Figure 1. Mutations to GmNOD26 alter water and cation permeability. (A) Osmotic water permeability; or (B) Na+ induced clamp currents; of control, Phosphomimetics or Wt GmNOD26-expressing X. laevis oocytes (n = 8).
References:
Byrt, C., Zhao, M., Kourghi, M., Bose, J., Henderson, S., Qiu, J., Gilliham, M., Schultz, C., Schwarz, M., Ramesh, S., Yool, A., and Tyerman, S. (2017) Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant, Cell & Environment, 40:802– 815.
Tyerman, S., Whitehead, L., and Day, D. (1995). A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature, 378:629–632.
Yool, A., Stamer, W., and Regan, J. (1996). Forskolin stimulation of water and cation permeability in aquaporin-1 water channels. Science, 273:1216–1218.
Miss Yuhan Liu, Ph.D. candidate | Wonder of Science Young Science Ambassador, School of Agriculture & Food Sciences | Integrative Legume Research Group, Faculty of Science | The University of Queensland.
What is the title of your poster?
Root meristem growth factor peptides and their effects on root development and nodulation in soybean

What is the aim of your research, and what is the main hypothesis that you are testing?
The research aims to understand the role of a new peptide family, RGFs, in legume growth. Given its orthologous to Arabidopsis and Medicago peptides regulating meristem activity and immune response, we hypothesized that these RGF peptides affect root growth and nodule development in soybean.
What plant species was the focus of your research, and what were the key techniques used?
Soybean (Glycine max). A range of molecular and physiological techniques were used to characterise gene function, including Agrobacterium-mediated Hairy Root transformation, gene overexpression, promotor::GUS fusions, microscopy, bioinformatics and phenotypic analyses.
What was the major finding(s) of your research?
A number of GmRGFs are specifically expressed in soybean nodules and root tips. They are critical for nodule maturation.
Who were the important collaborators and funders of your work?
Members of the Integrative Legume Research Group, the University of Queensland were important to help carry out this research project. Xitong Chu from The University of Queensland and Huazhong Agricultural University. Funding for April Hastwell and Brett Ferguson is provided by the Australian Research Council (ARC; DE200100800 and DP190102996). Yuhan Liu is supported by a University of Queensland Earmarked Scholarship, a UQ School of Agriculture and Food Sciences Travel Award and a ASPS Student Travel Award.
Recent Posts
Tags
Archives
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- February 2024
- January 2024
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
Copyright 2017 Australian Society of Plant Scientists Disclaimer & Privacy
Website by Michael Major Media


